
The role of molecular pathology in mediastinal sarcomas
Introduction
Primary mediastinal sarcomas represent an extremely rare and heterogenous group of mesenchymal neoplasms comprising <10% of primary mediastinal tumors and 1–2% of all soft tissue sarcomas (1-4). Despite their rare occurrence, primary mediastinal sarcomas tend to have an aggressive clinical course with a poor overall survival, usually worse than that of other types of mediastinal tumors (4-6). The mediastinal compartment is unique in that it houses multiple different tissue types allowing for great heterogeneity of both benign and malignant mesenchymal tumor types (7,8). It is difficult to determine the exact incidence of sarcoma subtypes arising within the mediastinum given their rarity and because multiple studies have reported different incidences for the different tumor types (4-7,9,10). Although predicated upon the accuracy of the initial diagnosis and proper reporting, evaluation of the National Cancer Database and the Surveillance, Epidemiology, and End Results Program (SEER) database highlights a group of tumors that appear to occur most commonly as primary mediastinal sarcomas (5,9). These tumors include synovial sarcoma, liposarcoma, malignant peripheral nerve sheath tumor (MPNST), small round blue cell sarcomas (including Ewing sarcoma) and leiomyosarcoma (LMS) (4-7,9). A single study lists angiosarcoma with the common mediastinal sarcomas, however it is unclear whether these represented primary cardiac angiosarcomas or true primary mediastinal angiosarcomas (5). A wide variety of other sarcoma types have also been reported to occur in the mediastinum (Table 1) (6,8,9,11-24). Another soft tissue mesenchymal neoplasm that occurs with some frequency within the mediastinum is solitary fibrous tumor (SFT). Although generally regarded as having indolent or low-grade behavior, these tumors may behave in an aggressive manner and have a small, but definitive, malignant potential and are therefore included in this discussion (25,26). In addition, a recently described entity that can occur within the mediastinum, pleura, or chest wall; SMARCA4-deficient undifferentiated tumor/sarcoma will be briefly discussed (27).

Full table
The differential diagnosis of mediastinal sarcomas may be difficult due to their rare nature and often overlapping clinical and histologic features. Generally, the workup and diagnosis of primary mediastinal sarcomas is similar to that of their extramediastinal soft tissue counterparts. Traditionally the diagnosis has been based on a combination of clinical, radiological, histologic and immunohistochemical features, however, various cytogenetic and molecular tests have proved to be valuable ancillary modalities to help differentiate soft tissue sarcomas (28-31). Within the past decade the emergence of massively parallel sequencing technologies such as next generation sequencing (NGS) have allowed for more comprehensive evaluation of soft tissue sarcomas and the discovery of new genetic driver events underlying these lesions (32-37). Although the management of sarcomas is still evolving, access to new information about the underlying molecular genetic events driving these lesions will soon become a routine part of clinical practice as personalized medicine continues to advance. This review aims to focus on molecular diagnostics and their contribution to the differential diagnosis of mediastinal sarcomas with a focus on the most common tumor subtypes.
General molecular features and use of molecular diagnostics in mediastinal sarcomas
Bone and soft tissue tumors of the mediastinum can generally be characterized into two broad categories: those with complex karyotypes that lack recurrent genomic alterations and those with relatively simple karyotypes that harbor specific, recurrent genomic alterations. Tumors with a high degree of genomic instability, such as LMS, osteosarcoma and undifferentiated pleomorphic sarcoma, generally do not lend themselves to molecular diagnostic testing due to their lack of specific or recurrent genetic aberrations, although investigations may still be carried out to rule out other sarcoma subtypes in the differential diagnosis or provide the maximal amount of information on the patient’s tumor to the clinician (such as identifying a potential alteration that would qualify a patient for an experimental trial). However, a large subgroup of bone and soft tissue tumors which occur within the mediastinum are characterized by consistent and recurrent genetic abnormalities similar to their extra-mediastinal counterparts. For this group of tumors, the high rate of specific, recurrent genetic abnormalities allows molecular testing to offer important diagnostic and prognostic information, help guide clinical management and treatment, and allows for recruitment of patients in ongoing clinical trials (37).
Genetic features of mediastinal sarcomas
Recurrent genetic abnormalities in soft tissue tumors in general most commonly fall into one of four categories: translocations, activating oncogenic mutations, inactivating oncogenic mutations, and copy number alterations such as gains, deletions, and amplifications (38). The commonest mediastinal sarcomas are almost exclusively comprised of tumors defined by translocations, inactivating oncogenic mutations, and amplification events of specific chromosomal regions (Table 2).

Full table
Translocations
Approximately 30% of bone and soft tissue tumors are characterized by recurrent chromosomal translocations (38-40). Translocations are rearrangements of genetic material that lead to novel juxtapositions of particular genes based on where chromosomal breaks have occurred. These gene rearrangements result in the formation of fusion oncogenes which are often involved in the pathogenesis and proliferation of the neoplastic cells, although it is worth noting that not all rearrangements lead to functional fusion proteins depending on the genes translocated as well the orientation of the genes themselves. One of the best characterized gene fusions in soft tissue sarcoma is the EWSR1 translocation in Ewing sarcoma. The most common rearrangement involves the long of arm of chromosome 11 with the long arm of chromosome 12, t(11;22)(q24q12), leading to a FLI1-EWSR1 fusion gene (41). At the molecular level fusions in many tumors may occur through a diverse combination of genetic material with various different transcripts existing for one fusion gene (see Ewing sarcoma section). It is worth noting that recurrent translocations usually occur in the background of simple karyotypes and that some genes involved in translocations have a diverse array of partner genes. In addition, certain genes are known to be involved in numerous translocations across multiple tumor types, for example, other than Ewing sarcoma the EWSR1 gene can also be identified in the translocations of round cell liposarcoma, extraskeletal myxoid chondrosarcoma, atypical Ewing sarcomas, and clear cell sarcoma amongst others (42).
Inactivating oncogenic mutations
Single nucleotide variants as well as small insertions and deletions are responsible for a variety of inactivation events in many different tumor types. In the mediastinum, a small group of sarcomas are characterized primarily by inactivating oncogenic events including extrarenal rhabdoid tumor, epithelioid sarcoma, epithelioid MPNST and the recently described SMARCA4-deficient undifferentiated thoracic tumor/sarcoma (1,4,17,20,27). Loss of gene function may occur through a combination of events including deletions, inactivating point mutations, copy neutral loss of heterozygosity and epigenetic events such as DNA methylation and histone modification (43-46). The diverse modes of gene inactivation present in these tumor types may require multiple testing modalities to accurately identify, although NGS with large mutation/fusion panels that also provide copy number information allow for evaluation of many of these changes in a single test (37,45,47).
Amplification events
Copy number alterations and amplification events occur less commonly in mediastinal sarcomas and usually apply to sarcomas which fall into the complex karyotype category of tumors (undifferentiated high-grade sarcomas, LMS and osteosarcoma). Well-differentiated and dedifferentiated liposarcoma are also characterized by a recurrent amplification event (48-51). Tumors in the high complexity category tend to have multiple different chromosomal events leading to copy number alterations in combination with other types of molecular abnormalities such as mutations—the majority of these tend to be non-recurrent events between different tumor subtypes as well as within the same tumor types making molecular evaluation of the lesions less useful in routine clinical practice. However, amplification events of a specific region of 12q13-15 tend to be the sole abnormality in well-differentiated liposarcoma and dedifferentiated liposarcoma (although de-differentiated may have additional genetic alterations such as mutations) (50,52). Copy number alterations in soft tissue neoplasms are particularly amenable to rapid diagnostic testing by fluorescence in situ hybridization (FISH) when probes are available, but may also be identified by array-based techniques or sequencing assays.
Advantages and disadvantages of different cytogenetic/molecular tests
As mentioned before, FISH, array-based assays including array comparative genomic hybridization (aCGH)/single nucleotide polymorphism (SNP) arrays, polymerase chain reaction (PCR) based assays and early-generation sequencing technologies such as Sanger sequencing and pyrosequencing have all traditionally constituted the backbone of molecular diagnostics in bone and soft tissue tumors (29,30,32). More recently the introduction of massively parallel targeted sequencing assays, including NGS, has opened new avenues for testing of soft tissues tumors by allowing a single tumor to be tested for multiple different genetic abnormalities with one assay (35-37). All of these technologies have certain advantages and disadvantages (Table 3) and the type of testing offered by different laboratories varies widely making it important for pathologists to understand the advantages and limitations of particular assays to identify the proper type of test for a identifying a particular genetic alteration. For example, FISH and PCR may be used to identify specific genetic abnormalities in a case where a particular diagnosis is suspected at a low price with a rapid turnaround time. While array, karyotype and sequencing may be better options for difficult tumors where the diagnosis is unknown and the pathologist desires the maximum amount of genetic information to help inform the differential diagnosis. Particular attention should also be paid to the type of sample required for the different assays with the caveat that many molecular assays such as FISH, array, PCR, and NGS may not work properly or completely fail when attempted on decalcified specimens due to the degradation of nucleic acids (53,54).
Full table
Common mediastinal sarcomas
While primary mediastinal sarcomas are extremely rare tumors in general, there is a group of sarcomas that appear to occur more commonly across multiple published series as compared to other sarcoma subtypes (1,3,5,6,8,9). These mediastinal sarcomas for the most part have some specific, recurrent genetic abnormalities that allow for molecular testing to play a significant role in the diagnosis of these tumors. It is important to note that many of the lesions in this category present with a characteristic clinical picture, as well as distinct morphological and immunohistochemical profiles that potentially allows for diagnosis without molecular diagnostics. However, molecular testing can help confirm a suspected diagnosis and is also useful for high grade lesions where the morphology or immunohistochemistry (IHC) profile may be non-specific. As mentioned previously, the mediastinum encompasses multiple different tissues types and depending on one’s definition of the mediastinal compartment the diagnosis of a primary mediastinal sarcoma may change. For example, true primary mediastinal angiosarcoma unassociated with the heart is exceedingly rare (55), despite being included in the analysis of some studies (5). What follows is a brief review of the molecular aspects of the most common mediastinal sarcomas.
Synovial sarcoma
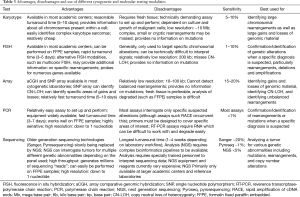
Synovial sarcoma has been traditionally grouped with tumors of uncertain histogenesis. Recent molecular expression studies suggest a myogenic origin, although this is inconsistent with the characteristic IHC profile that shows epithelial marker expression and lack of expression of myogenic markers (56). Synovial sarcoma is defined by a fusion oncoprotein; SS18-SSX, resulting from a characteristic reciprocal t(X;18)(p11.2;q11.2). This translocation fuses SS18 with one of three SSX genes clustered on chromosome X (Xp11.2); either SSX1, SSX2, or rarely SSX4 (57). An alternative, much less common, t(X;20)(p11;q13) has also been described which leads to an SS18L1-SSX1 fusion oncoprotein (58). The fusion oncoprotein results in dysregulation of SS18, a member of the chromatin remodeling SWI/SNF (BAF) complex (56). Studies correlating specific SS18-SSX fusion transcripts with histologic subtype or impact on prognosis have had conflicting results and primary mediastinal synovial sarcomas have not been well represented (59-61). Diagnosis of the fusion protein may be quickly performed with reasonably high sensitivity by FISH and PCR (Figure 1) (59,60). Testing of synovial sarcoma with NGS fusion panels allows for identification of the specific partner genes which cannot be identified through FISH and may be missed by PCR depending on the primer sets or type of PCR assay used (57,62). Studies that have examined primary mediastinal synovial sarcomas have identified that they share the same genetic alterations as their soft tissue counterparts (63,64).
Liposarcoma
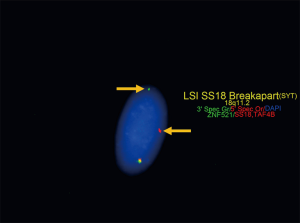
Liposarcoma is a tumor of adipocytic origin characterized by supernumerary ring and marker chromosomes (Figure 2A). The supernumerary chromosomes are most often composed of genomic material from the q13-15 region on chromosome 12 (65,66). The q13-15 region contains several genes, including the genes MDM2 and CDK4 (67). Although many other genetic alterations including numerous somatic mutations, other gene amplification events, and gene deletions have been described in well- and dedifferentiated liposarcoma, amplification of MDM2 and CDK4 (particularly MDM2) has been accepted as the diagnostic criteria for well-differentiated liposarcoma and de-differentiated liposarcoma (68-70). MDM2 amplification leads to overexpression of MDM2, promoting tumorigenesis by dysregulation of the p53 pathway (Figure 2B) (71). Amplification of CDK4 leads to overexpression of CDK4 that can then bind to cyclin D at increased levels, leading to interference with the E2F-RB interaction that acts as a cell cycle progression check from the G1 to S phase transition (72). Few series exist examining mediastinal liposarcomas; however, the predominant tumor subtypes across all series appear to be well- and dedifferentiated liposarcoma, the majority of which showed the characteristic amplifications or IHC expression (73-76). Amplification of MDM2 and CDK4 has been shown to correlate extremely well with IHC for MDM2 and CDK4, allowing IHC to serve as an excellent screening tool, however confirmation of the diagnosis by molecular techniques is always recommended (77). Generally, this is best accomplished with FISH using probes specific to MDM2 that are widely available, although copy number alterations may be assessed by array as well as certain NGS-based assays that provide copy number information (70). Of note, other subtypes of liposarcoma, such as myxoid and pleomorphic, may less commonly occur in the mediastinum (75).
LMS
LMS represents a mesenchymal tumor of smooth muscle origin characterized by complex cytogenetic and molecular aberrations (78,79). Cytogenetically LMS displays complex karyotypic changes that can include numerous gains, losses, rearrangements as well as chromothripsis (chromosomal shattering) in up to 35% of cases (79). Targeted exome sequencing studies have identified that the most frequent cytogenetic changes involve losses of material containing important tumor suppressor genes including PTEN (10q), RB1 (13q), CDH1 (16q), and TP53 (17p) (78). Mutations in TP53, RB1, and ATRX have been identified as the most common mutations to occur in LMS, although additional mutations in genes related to multiple signaling pathways, cell cycle regulation, DNA damage repair, muscle cell proliferation and epigenetic regulation are enriched as well (79). Primary mediastinal LMS have only been reported in small series or case reports, although they comprised a significant percentage of mediastinal sarcomas (~10%) in a large study which reviewed the National Cancer Database for cases of mediastinal sarcomas (5,80-82). Few, if any, primary mediastinal LMS’s have been examined by molecular techniques, although they likely share similar genetics to their soft tissue counterparts at extramediastinal sites. LMS is a clinicopathologic diagnosis and can often be made in the context of appropriate histomorphology and IHC supporting smooth muscle differentiation, making molecular genetic testing for the purposes of diagnosis less useful compared to sarcomas with specific, recurrent genetic abnormalities. However, clinically relevant molecular subtypes of LMS have been described and as new targeted therapies emerge molecular testing may play a larger role in guiding management and informing prognosis (83).
MPNST
MPNST is a tumor of neural origin which can occur in the mediastinum and which arises most commonly in patients with neurofibromas and neurofibromatosis type 1 (NF1) (5,9). The tumors occur most often within the posterior mediastinum in association with nerves located within that compartment, although rare cases have been described in the anterior mediastinum (84-87). These lesions may arise from malignant transformation of benign neural tumors, but may also occur sporadically (88,89). MPNST is characterized predominantly by recurrent mutations in NF1, TP53, CDKN2A, SUZ12, and EED (88-91). In particular, recurrent loss of function mutations in SUZ12 and EED lead to dysregulation of the polycomb repressive complex 2 (PRC2) with downstream dysregulation of the Ras pathway (92). MPNST’s with PRC2 loss have been shown to have loss of trimethylation at lysine 27 of histone H3 (H3K27me3) which can be identified with a monoclonal antibody against H3K27me3 (88,93). The epithelioid variant has been shown to have loss of SMARCB1 (94). The diagnosis of MPNST is currently accomplished primarily through clinicopathologic correlation and IHC, although the discovery of recurrent mutations in a high proportion of MPNST’s may allow molecular testing to play a larger role in their diagnosis.
Ewing sarcoma/primitive neuroectodermal tumor (PNET)
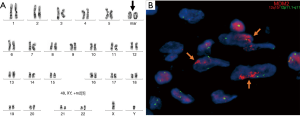
Ewing sarcoma represents an undifferentiated primitive small round blue cell sarcoma of uncertain histogenesis (8). This sarcoma is characterized by a classic t(11;22)(q24;q12) creating an oncogenic fusion protein; EWSR1-FLI1 (32,94). Less commonly, other EWSR1-ETS family rearrangements may occur (Table 2). The EWSR1-FLI1 fusion may form from various different transcripts at the molecular level; 60% are designated as type 1 fusions that fuse exon 7 of EWSR1 to exon 6 of FLI1, while 20% are designated as type 2 fusions that fuse exon 7 of EWSR1 to exon 5 of FLI1 (Figure 3A) (41). Numerous additional variant fusions have been described including breakpoints in the regions of exons 3 through 8 on FLI1, however these are less common than type 1 or type 2 changes (32). In past years the specific transcript identified held clinical significance as the patients could potentially respond to treatment differently, however, updated treatment regimens appear to have eliminated these differences and the need for reporting the exact transcript (41). Given that EWSR1 is a constant fusion partner in Ewing sarcoma, molecular testing can be rapidly and easily done using FISH break apart probes (Figure 3B) (95). As many of the EWSR1 fusion partners have been delineated over the years, reverse transcriptase PCR may also be used to accurately and quickly identify rearrangements (96). Tumors occurring within the mediastinum have been shown to harbor the characteristic translocations and are thus amenable to molecular testing for confirming the diagnosis (8,97,98). Of note, in recent years molecular diagnostics have expanded the spectrum of small round blue cell sarcomas (sometimes referred to as Ewing’s-like tumors) to include several new rearrangement partners for EWSR1 as well as other small round blue cell sarcomas with novel translocations such as CIC- and BCOR- rearranged sarcomas (99,100). Small round blue cell sarcomas that are negative for the classic Ewing’s translocation should be submitted for expanded molecular genetic testing to identify other possible translocations.
SMARCA4-deficient undifferentiated thoracic tumor/sarcoma
SMARCA4-deficient thoracic sarcoma (also known as SMARCA4-deficient undifferentiated tumor or SMARCA4-deficient thoracic sarcomatoid tumor) is a recently described entity of uncertain histogenesis (27). Some authors have postulated that these lesions represent undifferentiated epithelial malignancies and that they are part of the disease spectrum of SMARCA4-deficient carcinoma, particularly since many of the tumors studied have been identified to harbor smoking related genomic signatures (101). The tumors thus far designated as sarcomas tend to have different clinicopathologic parameters than the carcinomas, including different age ranges, extrapulmonary locations, and a different pattern of metastasis suggesting they represent a distinct clinicopathologic entity (27,102,103). A small percentage of these lesions also occur in the absence of a smoking history making their exact histogenesis unclear (101). Whether these undifferentiated tumors represent epithelial malignancies, mesenchymal malignancies or both still requires some additional clarification. While it remains unclear whether these tumors definitively arise as primary mediastinal sarcomas, the tumors can commonly involve the mediastinal compartment and some show no evidence of pulmonary involvement raising the possibility that some may indeed occur as primary mediastinal tumors (27,101-103). Given that a large percentage of these lesions involve the mediastinum they are included here as pathologists should include them in the differential diagnosis of poorly differentiated sarcomas with rhabdoid morphology.
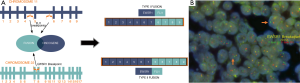
Molecularly these lesions are characterized by biallelic inactivation of the SMARCA4 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 4) gene that codes for Brahma-related gene 1 (BRG1); a member of the SWI/SNF complex (100,101). Inactivation occurs primarily through frameshift or nonsense mutations, although missense mutations, deletions, and splice site mutations have been identified as well (104). Mutations in TP53, NF1, KRAS, STK11, and KEAP1 may also occur in addition to the mutations in SMARCA4 (101,104). Cytogenetically the tumors can also show various copy number abnormalities, copy neutral loss of heterozygosity, and a generally complex genomic profile expected of a high-grade malignant neoplasm (104). Loss of SMARCA4 leads to dysregulation of the SWI/SNF (BAF) complex; a chromatin remodeling complex that is frequently mutated across many human cancers including synovial sarcoma, epithelioid MPNST, epithelioid sarcoma, and extrarenal rhabdoid tumor (Figure 4A) (105,106). Identification of the various mutations and complex genomic profiles of these tumors requires examination with advanced molecular techniques such as sequencing or a combination of various modalities (including array to identify copy neutral loss of heterozygosity), however the diagnosis can usually be made based on morphology combined with IHC showing loss of BRG1 expression (Figure 4B,C) (102,103).
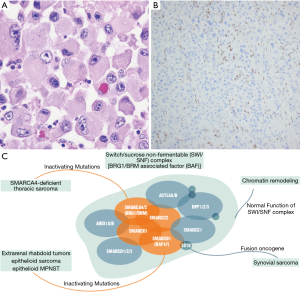
SFT
SFT is a fibroblastic mesenchymal tumor that can occur with some frequency within the structures of the thorax including the mediastinum (107-109). The tumor is characterized by a pathognomic NAB2-STAT6 fusion oncogene arising from a recurrent intrachromosomal rearrangement on chromosome 12q (109). Although most tumors follow an indolent clinical course, malignant variants or high-risk tumors have been reported, sometimes in association with additional genetic alterations such as TP53 and TERT promoter mutations (110-112). The fusion may be identified through sequencing, FISH, or reverse transcriptase PCR (107,108), however due to the close proximity of the genes involved in the translocation it may be missed by FISH. The diagnosis is also usually facilitated by IHC for STAT6 which is expressed in nearly 100% of tumors (111).
Less common mediastinal sarcomas
A wide diversity of other less common sarcoma subtypes have been reported to occur in the mediastinum as primary mediastinal tumors (Table 1) (6,8,9,11-24,113-115). These include sarcomas such as non-Ewing small round blue cell sarcomas and undifferentiated pleomorphic sarcomas, vascular tumors such as angiosarcoma and epithelioid hemangioendothelioma, bone tumors such as osteosarcoma, chondrosarcomas, and chordoma as well as various other more esoteric tumors such as follicular dendritic cell sarcoma, malignant PEComa, clear cell sarcoma and alveolar soft part sarcoma. Strict clinicopathologic correlation is required to rule out metastatic lesions from other primary soft tissue sites in these cases. Many of these lesions harbor recurrent or specific genetic abnormalities that can be identified through various molecular or cytogenetic techniques (Table 4). One such example is a less common variant of liposarcoma; myxoid liposarcoma. Unlike well-differentiated and dedifferentiated liposarcomas, myxoid liposarcomas follow a different oncogenic mechanism characterized by a t(12;16)(q13;p11) that leads to the formation of the fusion oncogenes, FUS-DDIT3 (95% of cases) or EWSR1-DDIT3 (5% of cases) (75). Despite these recurrent genetic abnormalities, it is worth noting that many lesions in the less common category may be diagnosed on clinicopathologic grounds, however the use of ancillary molecular testing to confirm diagnoses and help identify difficult lesions is always beneficial if available.
Full table
Conclusions
Mediastinal sarcomas represent a heterogenous group of rare tumors. There is a small subset that appears to occur more frequently compared to some of the less common lesions. Many of these mediastinal sarcomas have specific, recurrent genetic abnormalities that can be identified through various molecular techniques to aid in diagnosis. These genetic abnormalities are often similar to their extramediastinal bone and soft counterparts.
Acknowledgments
The author would like to thank Dana Green for providing graphic design for Figure 3B,C.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Mediastinum, for the series “Mediastinal Sarcomas”. The article has undergone external peer review.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/med-20-39). The series “Mediastinal Sarcomas” was commissioned by the editorial office without any funding or sponsorship. DIS served as the unpaid Guest Editor of the series.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gladish GW, Sabloff M, Munden R, et al. Primary thoracic sarcomas. Radiographics 2002;22:621-37. [Crossref] [PubMed]
- Shields TW, Robinson PG. Mesenchymal tumors of the mediastinum. In: Shields TW, LoCicero J, Ponn RB, et al. editors. General thoracic surgery. Philadelphia: Lippincott Williams & Wilkins, 2005:2786-800.
- Macchiarini P, Ostertag H. Uncommon primary mediastinal tumours. Lancet Oncol 2004;5:107-18. [Crossref] [PubMed]
- Paquette M, Truong P, Hart J, et al. Primary sarcoma of the mediastinum: a report of 16 cases referred to the British Columbia Cancer Agency. J Thorac Oncol 2010;5:898-906. [Crossref] [PubMed]
- Engelhardt KE, DeCamp MM, Yang AD, et al. Treatment approaches and outcomes for primary mediastinal sarcoma: analysis of 976 patients. Ann Thorac Surg 2018;106:333-9. [Crossref] [PubMed]
- Luo DX, Huang MJ, Xiong B, et al. Primary mediastinal sarcoma: surgical outcomes of 21 cases. Interact Cardiovasc Thorac Surg 2013;17:982-6. [Crossref] [PubMed]
- den Bakker MA, Marx A, Mukai K, et al. Mesenchymal tumours of the mediastinum - part I. Virchows Arch 2015;467:487-500. [Crossref] [PubMed]
- den Bakker MA, Marx A, Mukai K, et al. Mesenchymal tumours of the mediastinum - part II. Virchows Arch 2015;467:501-17. [Crossref] [PubMed]
- Abdel-Rahman O. An analysis of clinical characteristics and patient outcomes in primary mediastinal sarcomas. Expert Rev Anticancer Ther 2017;17:1071-6. [Crossref] [PubMed]
- Turbendian H, Seastedt KP, Shavladze N, et al. Extended resection of sarcomas involving the mediastinum: a 15-year experience. Eur J Cardiothorac Surg 2016;49:829-34. [Crossref] [PubMed]
- Venuta F, Pescarmona EO, Rendina EA, et al. Primary osteogenic sarcoma of the posterior mediastinum. Case report. Scand J Thorac Cardiovasc Surg 1993;27:169-73. [Crossref] [PubMed]
- Yu H, Wu Z, Cui Y, et al. Low-grade extraskeletal osteosarcoma of the mediastinum: report of a case and review of literature. Int J Clin Exp Pathol 2015;8:3279-81. [PubMed]
- Shimizu N, Tanaka Y, Demizu Y, et al. Surgery and proton beam therapy for mediastinal extraskeletal osteosarcoma. Ann Thorac Surg 2019;108:e289-91. [Crossref] [PubMed]
- Jin L, Sui Y, Zhu H, et al. Primary mediastinal clear cell sarcoma: a case report and review of the literature. Diagn Pathol 2017;12:5. [Crossref] [PubMed]
- Tirabosco R, Lang-Lazdunski L, Diss TC, et al. Clear cell sarcoma of the mediastinum. Ann Diagn Pathol 2009;13:197-200. [Crossref] [PubMed]
- Maeda E, Ohta S, Watadani T, et al. Imaging findings of thoracic low-grade fibromyxoid sarcoma: report of three cases. Jpn J Radiol 2009;27:375-80. [Crossref] [PubMed]
- Gross E, Rao BN, Pappo A, et al. Epithelioid sarcoma in children. J Pediatr Surg 1996;31:1663-5. [Crossref] [PubMed]
- Flieder DB, Moran CA, Suster S. Primary alveolar soft-part sarcoma of the mediastinum: a clinicopathological and immunohistochemical study of two cases. Histopathology 1997;31:469-73. [Crossref] [PubMed]
- Suster S, Moran CA. Malignant cartilaginous tumors of the mediastinum: clinicopathologic study of six cases presenting as extraskeletal soft tissue masses. Hum Pathol 1997;28:588-94. [Crossref] [PubMed]
- Parham DM, Weeks DA, Beckwith JB. The clinicopathologic spectrum of putative extrarenal rhabdoid tumors. An analysis of 42 cases studied with immunohistochemistry or electron microscopy. Am J Surg Pathol 1994;18:1010-29. [Crossref] [PubMed]
- Liang W, Xu S, Chen F. Malignant perivascular epithelioid cell neoplasm of the mediastinum and the lung. Medicine (Baltimore) 2015;94:e904 [Crossref] [PubMed]
- Leipsic JA, McAdams HP, Sporn T. Follicular dendritic cell sarcoma of the mediastinum. AJR Am J Roentgenol 2007;188:W554-6 [Crossref] [PubMed]
- Chow SC, Yeung EC, Ng CS, et al. Mediastinal follicular dendritic cell sarcoma with paraneoplastic pemphigus. Asian Cardiovasc Thorac Ann 2015;23:732-4. [Crossref] [PubMed]
- Spatola C, Migliore M, Liardo R, et al. Follicular dendritic cell sarcoma of mediastinum: a key role of radiotherapy in a multidisciplinary approach. Future Oncol 2015;11:57-61. [Crossref] [PubMed]
- Milano MT, Singh DP, Zhang H. Thoracic malignant solitary fibrous tumors: A population-based study of survival. J Thorac Dis 2011;3:99-104. [PubMed]
- Ronchi A, Cozzolino I, Marino F, et al. Extrapleural solitary fibrous tumor: a distinct entity from pleural solitary fibrous tumor. An update on clinical, molecular and diagnostic features. Ann Diagn Pathol 2018;34:142-50. [Crossref] [PubMed]
- Sauter JL, Graham RP, Larsen BT, et al. SMARCA4-deficient thoracic sarcoma: a distinctive clinicopathological entity with undifferentiated rhabdoid morphology and aggressive behavior. Mod Pathol 2017;30:1422-32. [Crossref] [PubMed]
- Thway K, Jordan S, Fisher C, et al. Updates in the approach to intrathoracic sarcomas. Histopathology 2015;67:755-70. [Crossref] [PubMed]
- Norberg SM, Movva S. Role of genetic and molecular profiling in sarcomas. Curr Treat Options Oncol 2015;16:24. [Crossref] [PubMed]
- Osuna D, de Alva E. Molecular pathology of sarcomas. Rev Recent Clin Trials 2009;4:12-26. [Crossref] [PubMed]
- Ordóñez JL, Osuna D, Garcia-Dominguez DJ, et al. The clinical relevance of molecular genetics in soft tissue sarcomas. Adv Anat Pathol 2010;17:162-81. [Crossref] [PubMed]
- Bridge JA, Cushman-Vokoun AM. Molecular diagnostics of soft tissue tumors. Arch Pathol Lab Med 2011;135:588-601. [PubMed]
- Torres-Mora J, Fritchie KJ, Robinson SI, et al. Molecular genetics of soft-tissue sarcomas: a brief overview for clinical oncologists. Cancer J 2014;20:73-9. [Crossref] [PubMed]
- Szurian K, Kashofer K, Liegl-Atzwanger B. Role of next-generation sequencing as a diagnostic tool for the evaluation of bone and soft-tissue tumors. Pathobiology 2017;84:323-38. [Crossref] [PubMed]
- Baumhoer D, Amary F, Flanagan AM. An update of molecular pathology of bone tumors. Lessons learned from investigating samples by next generation sequencing. Genes Chromosomes Cancer 2019;58:88-99. [Crossref] [PubMed]
- Lam SW, Cleton-Jansen AM, Cleven AGH, et al. Molecular analysis of gene fusions in bone and soft tissue tumors by anchored multiplex PCR-based targeted next-generation sequencing. J Mol Diagn 2018;20:653-63. [Crossref] [PubMed]
- Cote GM, He J, Choy E. Next-generation sequencing for patients with sarcoma: A single center experience. Oncologist 2018;23:234-42. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive and Integrated Genomic Characterization of Adult Soft Tissue Sarcomas. Cell 2017;171:950-965.e28. [Crossref] [PubMed]
- Bridge JA, Nelson M. Genetics of soft tissue. In: Miettinen M. editor. Modern Soft Tissue Pathology Tumors and Non-Neoplastic Conditions. Washington, DC: Cambridge University Press, 2010:105-26.
- Nakano K, Takahashi S. Translocation-Related Sarcomas. Int J Mol Sci 2018;19:3784. [Crossref] [PubMed]
- van Doorninck JA, Ji L, Schaub B, et al. Current treatment protocols have eliminated the prognostic advantage of type 1 fusions in ewing sarcoma; a report from the children’s oncology group. J Clin Oncol 2010;28:1989-94. [Crossref] [PubMed]
- Romeo S, Dei Tos AP. Soft tissue tumors associated with EWSR1 translocation. Virchows Arch 2010;456:219-34. [Crossref] [PubMed]
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res 2011;21:381-95. [Crossref] [PubMed]
- Morris LG, Chan TA. Therapeutic targeting of tumor suppressor genes. Cancer 2015;121:1357-68. [Crossref] [PubMed]
- Jackson EM, Sievert A, Gai X, et al. Genomic analysis using high-density single nucleotide polymorphism-based oligonucleotide arrays and multiplex ligation-dependent probe amplification provides a comprehensive analysis of INI1/SMARCB1 in malignant rhabdoid tumors. Clin Cancer Res 2009;15:1923-30. [Crossref] [PubMed]
- Modena P, Lualdi E, Facchinetti F, et al. SMARCB1/INI1 tumor suppressor gene is frequently inactivated in epithelioid sarcomas. Cancer Res 2005;65:4012-19. [Crossref] [PubMed]
- Shen W, Szankasi P, Durtschi J, et al. Genome-wide copy number variation detection using NGS: data analysis and interpretation. Methods Mol Biol 2019;1908:113-24. [Crossref] [PubMed]
- Tarkkanen M, Karhu R, Kallioniemi A, et al. Gains and losses of DNA sequences in osteosarcomas by comparative genomic hybridization. Cancer Res 1995;55:1334-38. [PubMed]
- Boltze C, Schneider-Stock R, Jager V, et al. Distinction between lipoma and liposarcoma by MDM2 alterations: a case report of simultaneously occurring tumors and review of the literature. Pathol Res Pract 2001;197:563-8. [PubMed]
- Ware PL, Snow AN, Gvalani M, et al. MDM2 copy numbers in well-differentiated and dedifferentiated liposarcoma: characterizing progression to high-grade tumors. Am J Clin Path 2014;141:334-41. [Crossref] [PubMed]
- Kelleher FC, Viterbo A. Histologic and genetic advances in refining the diagnosis of “undifferentiated pleomorphic sarcoma”. Cancers (Basel) 2013;5:218-33. [Crossref] [PubMed]
- Thway K, Jones RL, Noujaim J, et al. Dedifferentiated liposarcoma: updates on morphology, genetics, and therapeutic strategies. Adv Anat Pathol 2016;23:30-40. [Crossref] [PubMed]
- Choi SE, Hong SW, Yoon SO. Proposal of an appropriate decalcification method of bone marrow biopsy specimens in the era of expanding genetic molecular study. J Pathol Transl Med 2015;49:236-42. [Crossref] [PubMed]
- Singh VM, Salunga RC, Huang VJ, et al. Analysis of the effect of various decalcification agents on the quantity and quality of nucleic acid (DNA and RNA) recovered from bone biopsies. Ann Diagn Pathol 2013;17:322-6. [Crossref] [PubMed]
- Weissferdt A, Kalhor N, Suster S, et al. Primary angiosarcomas of the anterior mediastinum: a clinicopathologic and immunohistocemical study of 9 cases. Hum Path 2010;41:1711-7. [Crossref] [PubMed]
- McBride MJ, Pulice JL, Beird HC, et al. The SS18-SSX Fusion Oncoprotein Hijacks BAF Complex Targeting and Function to Drive Synovial Sarcoma. Cancer Cell 2018;33:1128-1141.e7. [Crossref] [PubMed]
- Charlson J, Bedi M, Suster D, et al. Synovial Sarcoma; In: Trent JC, Delaney T, Pollock R, et al. editors. Sarcomas: Evidence-Based Diagnosis and Management, 1st edition. New York, New York: Springer Publishing Company, 2019.
- Storlazzi CT, Mertens F, Mandahl N, et al. A novel fusion gene, SS18L1/SSX1, in synovial sarcoma. Genes Chromosomes Cancer 2003;37:195-200. [Crossref] [PubMed]
- Amary MF, Berisha F, Bernardi Fdel C, et al. Detection of SS18-SSX fusion transcripts in formalin-fixed paraffin-embedded neoplasms: analysis of conventional RT-PCR, qRT-PCR and dual color FISH as diagnostic tools for synovial sarcoma. Mod Pathol 2007;20:482-96. [Crossref] [PubMed]
- Shipley J, Crew J, Birdsall S, et al. Interphase fluorescence in situ hybridization and reverse transcription polymerase chain reaction as a diagnostic aid for synovial sarcoma. Am J Pathol 1996;148:559-67. [PubMed]
- dos Santos NR, de Bruijin DR, van Kessel AG. Molecular mechanisms underlying human synovial sarcoma development. Genes Chromosomes Cancer 2001;30:1-14. [Crossref] [PubMed]
- Zhu G. Diagnosis of known sarcoma fusions and novel fusion partners by targeted RNA sequencing with identification of a recurrent ACTB-FOSB fusion in pseudomyogenic hemangioendothelioma. Mod Pathol 2019;32:609-20. [Crossref] [PubMed]
- Lan T, Chen H, Xiong B, et al. Primary pleuropulmonary and mediastinal synovial sarcoma: a clinicopathologic and molecular study of 26 genetically confirmed cases in the largest institution of southwest China. Diagn Pathol 2016;11:62. [Crossref] [PubMed]
- Terra SBSP, Aesif SW, Maleszewski JJ, et al. Mediastinal Synovial Sarcoma: Clinicopathologic Analysis of 21 Cases With Molecular Confirmation. Am J Surg Pathol 2018;42:761-6. [Crossref] [PubMed]
- Micci F, Teixeira MR, Bjerkehagen B, et al. Characterization of supernumerary rings and giant marker chromosomes in well-differentiated lipomatous tumors by a combination of G-banding, CGH, M-FISH, and chromosome- and locus-Specific FISH. Cytogenet Genome Res 2002;97:13-9. [Crossref] [PubMed]
- Yang L, Chen S, Luo P, et al. Liposarcoma: advances in cellular and molecular genetics alterations and corresponding clinical treatment. J Cancer 2020;11:100-7. [Crossref] [PubMed]
- Pedeutour F, Forus A, Coindre JM, et al. Structure of the supernumerary ring and giant rod chromosomes in adipose tissue tumors. Genes Chromosomes Cancer 1999;24:30-41. [Crossref] [PubMed]
- Italiano A, Bianchini L, Gjernes E, et al. Clinical and Biological Significance of CDK4 Amplification in Well-Differentiated and Dedifferentiated Liposarcomas. Clin Cancer Res 2009;15:5696-703. [Crossref] [PubMed]
- Taylor BS, DeCarolis PL, Angeles CV, et al. Frequent alterations and epigenetic silencing of differentiation pathway genes in structurally rearranged liposarcomas. Cancer Discov 2011;1:587-97. [Crossref] [PubMed]
- Kanojia D, Nagata Y, Garg M, et al. Genomic landscape of liposarcoma. Oncotarget 2015;6:42429-44. [Crossref] [PubMed]
- Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer 2006;6:909-23. [Crossref] [PubMed]
- Shim BY, Yoo J, Lee YS, et al. Prognostic role of Rb, p16, Cyclin D1 proteins in soft tissue sarcomas. Cancer Res Treat 2010;42:144-50. [Crossref] [PubMed]
- Hahn HP, Fletcher CD. Primary mediastinal liposarcoma: clinicopathologic analysis of 24 cases. Am J Surg Pathol 2007;31:1868-74. [Crossref] [PubMed]
- Boland JM, Colby TV, Folpe AL. Liposarcomas of the mediastinum and thorax: a clinicopathologic and molecular cytogenetic study of 24 cases, emphasizing unusual and diverse histologic features. Am J Surg Pathol 2012;36:1395-403. [Crossref] [PubMed]
- Ortega P, Suster D, Falconieri G, et al. Liposarcomas of the posterior mediastinum: clinicopathologic study of 18 cases. Mod Pathol 2015;28:721-31. [Crossref] [PubMed]
- Miura K, Hamanaka K, Matsuoka S, et al. Primary mediastinal dedifferentiated liposarcoma: Five case reports and a review. Thorac Cancer 2018;9:1733-40. [Crossref] [PubMed]
- Aleixo PB, Hartmann AA, Menezes IC, et al. Can MDM2 and CDK4 make the diagnosis of well differentiated/dedifferentiated liposarcoma? An immunohistochemical study on 129 soft tissue tumours. J Clin Pathol 2009;62:1127-35. [Crossref] [PubMed]
- Agaram NP, Zhang L, LeLoarer F, et al. Targeted exome sequencing profiles genetic alterations in leiomyosarcoma. Genes Chromosomes Cancer 2016;55:124-30. [Crossref] [PubMed]
- Chudasama P, Mughal SS, Sanders MA, et al. Integrative genomic and transcriptomic analysis of leiomyosarcoma. Nat Commun 2018;9:144. [Crossref] [PubMed]
- Moran CA, Suster S, Perino G, et al. Malignant smooth muscle tumors presenting as mediastinal soft tissue masses. A clinicopathologic study of 10 cases. Cancer 1994;74:2251-60. [Crossref] [PubMed]
- Vaziri M. Primary mediastinal leiomyosarcoma. Gen Thorac Cardiovasc Surg 2012;60:522-4. [Crossref] [PubMed]
- Xue X, Liang W, Zhang W. Anterior mediastinal leiomyosarcoma mimicking thymoma: A case report. Medicine (Baltimore) 2018;97:e11132 [Crossref] [PubMed]
- Guo X, Jo VY, Mills AM, et al. Clinically Relevant Molecular Subtypes in Leiomyosarcoma. Clin Cancer Res 2015;21:3501-11. [Crossref] [PubMed]
- Kamran SC, Shinagare AB, Howard SA, et al. Intrathoracic malignant peripheral nerve sheath tumors: imaging features and implications for management. Radiol Oncol 2013;47:230-8. [Crossref] [PubMed]
- Kalra B, Kingsley PA, Bedi HS, et al. Malignant peripheral nerve sheath tumor of the anterior mediastinum: a rare presentation. Rare Tumors 2014;6:5528. [Crossref] [PubMed]
- Nakajima Y, Mikami I, Akiyama H, et al. Long-term progress of six cases of malignant peripheral nerve sheath tumors of the mediastinum that underwent surgical treatment: Case report series. Int J Surg Case Rep 2016;24:185-7. [Crossref] [PubMed]
- Koezuka S, Hata Y, Sato F, et al. Malignant peripheral nerve sheath tumor in the anterior mediastinum: A case report. Mol Clin Oncol 2014;2:987-90. [Crossref] [PubMed]
- Lee W, Teckie S, Wiesner T, et al. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat Genet 2014;46:1227-32. [Crossref] [PubMed]
- Sohier P, Luscan A, Lloid A, et al. Confirmation of mutation landscape of NF1-associated malignant peripheral nerve sheath tumors. Genes Chromosomes Cancer 2017;56:421-6. [Crossref] [PubMed]
- Verdijk RM, den Bakker MA, Dubbink HJ, et al. TP53 mutation analysis of malignant peripheral nerve sheath tumors. J Neuropathol Exp Neurol 2010;69:16-26. [Crossref] [PubMed]
- Brohl AS, Kahen E, Yoder SJ, et al. The genomic landscape of malignant peripheral nerve sheath tumors: diverse drivers of Ras pathway activation. Sci Rep 2017;7:14992. [Crossref] [PubMed]
- De Raedt T, Beert E, Pasmant E, et al. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature 2014;514:247-51. [Crossref] [PubMed]
- Prieto-Granada CN, Wiesner T, Messina JL, et al. Loss of H3K27me3 expression is a highly sensitive marker for sporadic and radiation-induced MPNST. Am J Surg Pathol 2016;40:479-89. [Crossref] [PubMed]
- Jo VY, Fletcher CDM. Epithelioid malignant peripheral nerve sheath tumor: clinicopathologic analysis of 63 cases. Am J Surg Pathol 2015;39:673-82. [Crossref] [PubMed]
- Baldauf MC, Orth MF, Dallmayer M, et al. Robust diagnosis of Ewing sarcoma by immunohistochemical detection of super-enhancer-driven EWSR1-ETS targets. Oncotarget 2017;9:1587-601. [Crossref] [PubMed]
- Machado I, Navarro L, Pellin A, et al. Defining Ewing and Ewing-like small round cell tumors (SRCT): The need for molecular techniques in their categorization and differential diagnosis. A study of 200 cases. Ann Diagn Pathol 2016;22:25-32. [Crossref] [PubMed]
- Manduch M, Dexter DF, Ellis PM, et al. Extraskeletal Ewing's sarcoma/primitive neuroectodermal tumor of the posterior mediastinum with t(11;22)(q24;q12). Tumori 2008;94:888-91. [Crossref] [PubMed]
- Halliday J, Soon SY, Monaghan H, et al. Extraskeletal Ewing's sarcoma presenting as a mediastinal mass. Ann Thorac Surg 2010;90:1016-17. [Crossref] [PubMed]
- Pierron G, Tirode F, Lucchesi C, et al. A new subtype of bone sarcoma defined BCOR-CCNB3 gene fusion. Nat Genet 2012;44:461-6. [Crossref] [PubMed]
- Kawamura-Saito M, Yamazaki Y, Kaneko K, et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum Mol Genet 2006;15:2125-37. [Crossref] [PubMed]
- Rekhtman N, Montecalvo J, Chang JC, et al. SMARCA4-Deficient thoracic sarcomatoid tumors represent primarily smoking-related undifferentiated carcinomas rather than primary thoracic sarcomas. J Thorac Oncol 2020;15:231-47. [Crossref] [PubMed]
- Perret R, Chalabreysse L, Watson S, et al. SMARCA4-deficient thoracic sarcomas: clinicopathologic study of 30 cases with an emphasis on their nosology and differential diagnoses. Am J Surg Pathol 2019;43:455-5. [Crossref] [PubMed]
- Yoshida A, Kobayashi E, Kubo T, et al. Clinicopathological and molecular characterization of SMARCA4-deficient thoracic sarcomas with comparison to potentially related entities. Mod Pathol 2017;30:797-809. [Crossref] [PubMed]
- Le Loarer F, Watson S, Pierron G, et al. SMARCA4 inactivation defines a group of undifferentiated thoracic malignancies transcriptionally related to BAF-deficient sarcomas. Nat Genet 2015;47:1200-5. [Crossref] [PubMed]
- Agaimy A, Foulkes WD. Hereditary SWI/SNF complex deficiency syndromes. Semin Diagn Pathol 2018;35:193-8. [Crossref] [PubMed]
- Wang X, Haswell JR, Roberts CW. Molecular pathways: SWI/SNF (BAF) complexes are frequently mutated in cancer—mechanisms and potential therapeutic insights. Clin Cancer Res 2014;20:21-7. [Crossref] [PubMed]
- Suster S, Nascimento AG, Miettinen M, et al. Solitary fibrous tumors of soft tissue. A clinicopathologic and immunohistochemical study of 12 cases. Am J Surg Pathol 1995;19:1257-66. [Crossref] [PubMed]
- Akaike K, Kurisaki-Arakawa A, Hara K, et al. Distinct clinicopathological features of NAB2-STAT6 fusion gene variants in solitary fibrous tumor with emphasis on the acquisition of highly malignant potential. Hum Pathol 2015;46:347-56. [Crossref] [PubMed]
- Ghanim B, Hess S, Bertoglio P, et al. Intrathoracic solitary fibrous tumor - an international multicenter study on clinical outcome and novel circulating biomarkers. Sci Rep 2017;7:12557. [Crossref] [PubMed]
- Bahrami A, Lee S, Schaefer IM, et al. TERT promoter mutations and prognosis in solitary fibrous tumor. Mod Pathol. 2016;29:1511-22. [Crossref] [PubMed]
- Thway K, Ng W, Noujaim J, et al. The current status of solitary fibrous tumor: diagnostic features, variants, and genetics. Int J Surg Pathol 2016;24:281-92. [Crossref] [PubMed]
- De Raet J, Sacré R, Hoorens A, et al. Malignant giant solitary fibrous tumor of the mediastinum. J Thorac Oncol 2008;3:1068-70. [Crossref] [PubMed]
- Okuda K, Yano M, Moriyama S, et al. A case of mediastinum undifferentiated high grade pleomorphic sarcoma. Int J Clin Exp Med 2015;8:19566-70. [PubMed]
- Suster S, Moran CA. Chordomas of the mediastinum: clinicopathologic, immunohistochemical, and ultrastructural study of six cases presenting as posterior mediastinal masses. Hum Pathol 1995;26:1354-62. [Crossref] [PubMed]
- Patrini D, Scolamiero L, Khiroya R, et al. Mediastinal hemangioendothelioma: Case report and review of the literature. Respir Med Case Rep 2017;22:19-23. [Crossref] [PubMed]
Cite this article as: Suster DI. The role of molecular pathology in mediastinal sarcomas. Mediastinum 2020;4:33.


